Computer-Aided Drug Design Methods PMC
Table Of Content
The quality of QSAR models, however, differs for different target classes depending on data availability, with the most advances achieved for the kinase superfamily and aminergic GPCRs. An unbiased benchmark of the best ML QSAR models was given by a recent IDG-DREAM Drug-Kinase Binding Prediction Challenge with the participation of more than 200 experts78. The top predictive models in this blind assessment included kernel learning, gradient boosting and DL-based algorithms. The top-performing model (from team Q.E.D) used a kernel regression, protein sequence similarity and affinity values of more than 60,000 compound–kinase pairs between 13,608 compounds and 527 kinases from ChEMBL79 and Drug Target Commons80 databases as the training data. The best DL model used as many as 900,000 experimental ligand-binding data points for training, but still trailed the much simpler kernel model in performance.
4 Docking-based VS
However, building, maintaining and searching fully enumerated chemical libraries comprising more than a few billion compounds become slow and impractical. Such gigascale virtual libraries are therefore usually maintained as non-enumerated chemical spaces, defined by a specific set of building blocks and reactions (or transforms), as comprehensively reviewed in ref. 38. Within pharma, one of the first published examples includes PGVL by Pfizer37,43, the most recent version of which uses a set of 1,244 reactions and in-house reagents to account for 1014 compounds. Other biopharma companies have their own virtual chemical spaces38,44, although their details are often not in the public domain. Among commercially available chemical spaces, GalaXi Space by WuXi (approximately 8 billion compounds), CHEMriya by Otava (11.8 billion compounds) and Enamine REAL Space (36 billion compounds)45 are among the largest and most established.
Ligand-based
The pharmacophore model generated should have optimum sensitivity and specificity to minimize the chances of false negative and false positive results and must be validated using an independent external test set [81]. If the information about the 3D structure of a receptor and a set of known active compounds are lacking, then a sequence-derived 3D pharmacophore model is quite useful. For example, Pharma3D utilizes knowledge of the 3D crystal structures and homology models to derive the common sequence motif important for receptor-ligand biomolecular interactions in protein families [81, 82]. Running such data-rich computationally driven pipelines requires overarching data management tools for drug discovery, many of them being implemented in pharma and academic DDD centres146,147.
3. Pharmacophore Modelling
The process starts with the biological identification of a putative target to which ligand binding should lead to antimicrobial activity. In SDBB, the 3D structure of the target can be identified by X-ray crystallography or NMR or using homology modeling. LBDD is used in the absence of the target 3D structure with the central theme being the development of an SAR from which information on modification of the lead compound to improve activity can be obtained.
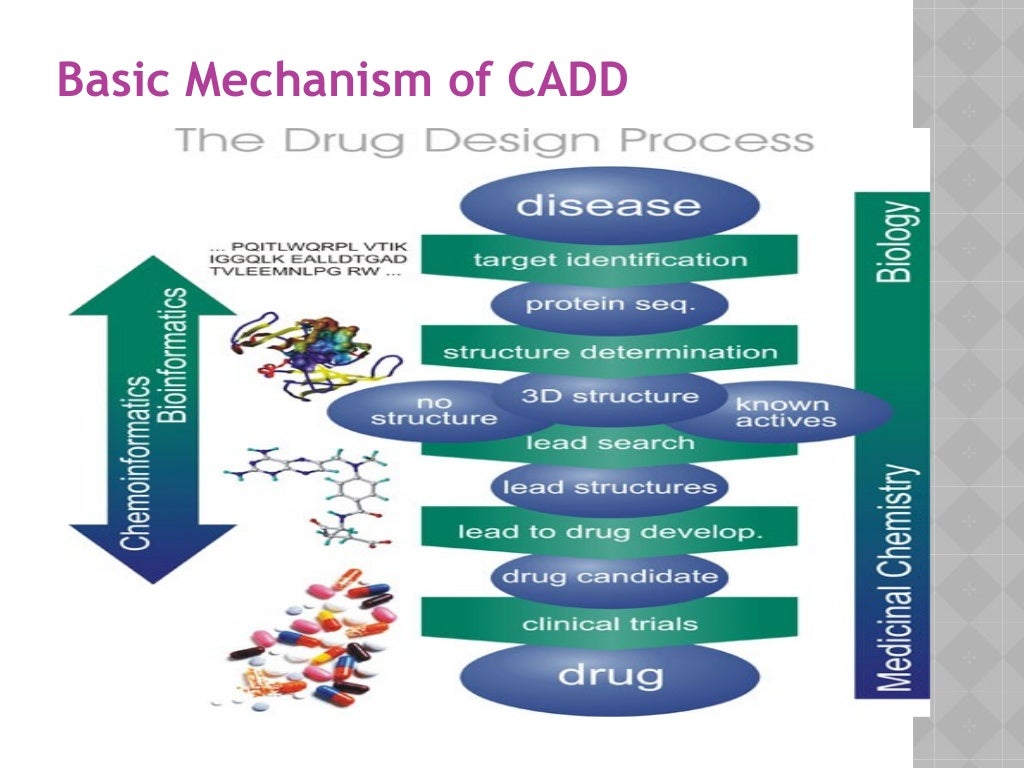
The applicability of CADD for modeling of the drug considers combinatorial chemistry and bioinformatics which address the major issues including cost and time duration. Computational binding site identification methods can be used to exploit novel, druggable sites on new protein targets for potential therapeutic development (113). For antibiotics development, such methods can be employed to search for putative allosteric sites as alternatives to the active or orthosteric sites on bacterial proteins to overcome drug resistance issues (114).
Pharmacophore models mapping
In collaborative studies with the Wilks lab, we have successfully applied CADD techniques to identify inhibitors of the bacterial heme oxygenases from Pseudomonas aeruginosa and Neisseria meningitides, thereby confirming the potential role of heme oxygenases as a novel antimicrobial targets (13, 14). A pharmacophore is an assembly (3D arrangement) of 'steric' and 'electronic' features required for optimal supramolecular interaction with a specific biological target structure and to prompt/block its biological response [63]. Ligand-based pharmacophore model generation is based on available information on the biological activities of compounds/ligands. A pharmacophore does not symbolize an actual molecule/ligand or real connection between functional groups, but rather provides an abstract description of molecular features that are vital for molecular interactions between molecules and macromolecular ligands. 3Protonation states of titratable residues at the targeted binding site and in the ligand being studied are quite important when setting up the CADD calculations.
A number of tools and software have been developed for pharmacophore development, such as, Phase, Catalyst/Discovery Studio, MOE, and LigandScout [64]. LBDD offers a general approach for elucidating relationships between the structural and physicochemical properties of compounds/ligands and their biological activities. In this process the available information of ligands and their biological activity is used for the development of new potential drug candidates. LBDD is widely used in pharmaceutical research, as more than 50% of approved drugs targeting membrane proteins (for which 3D structures are often not available, such as, GPCR).
Computer-Aided Drug Discovery Global Market Report 2022: Demand for Novel Drugs to Treat CVD Conditions Fuels ... - Yahoo Finance
Computer-Aided Drug Discovery Global Market Report 2022: Demand for Novel Drugs to Treat CVD Conditions Fuels ....
Posted: Mon, 07 Nov 2022 08:00:00 GMT [source]
This technique uses a known active compound as a query compound to find similar compounds and then rank compounds identified in a database. Based on this belief, structurally similar molecules exhibit similar biological activities and physicochemical properties. Numerical descriptors are applied and similarity coefficient is defined to quantify the degree of similarity (similarity/ dissimilarity).
4. Molecular Docking and Scoring Functions
Molecular docking has been greatly facilitated by dramatic growth in computer power and the increasing availability of small molecule and protein databases. Recent advancements in computer methods and access to 3D structural information of biological targets are set to increase the effectiveness of this technique and facilitate its large-scale application to studies of molecular interactions involved in ligand-protein binding. Many molecular docking programs have been developed during recent years, such as, AutoDock [40], Dock [41], FlexX [42], Glide [43], Gold [44], Surflex [45], ICM, and LigandFit [46], and been used successfully in many computer based drug discovery projects. Typically, the major goal of molecular docking is to identify ligands that bind most favorably within receptor binding sites and to determine its most energetically favored binding orientations (poses). A binding pose either refers to a conformation of a ligand molecule within the binding site of its target protein which has been confirmed experimentally, or a computationally modelled hypothetical conformation.
The pipeline involves the prediction of structure, visualization, binding site characterization, molecular docking and virtual screening, visualization of docked complex structure and their stability analysis, ADMET screening, and binding-free energy- MM PBSA will be involved. Computational approaches are useful tools to interpret and guide experiments to expedite the antibiotic drug design process. Structure based drug design (SBDD) and ligand based drug design (LBDD) are the two general types of computer-aided drug design (CADD) approaches in existence. SBDD methods analyze macromolecular target 3-dimensional structural information, typically of proteins or RNA, to identify key sites and interactions that are important for their respective biological functions. Such information can then be utilized to design antibiotic drugs that can compete with essential interactions involving the target and thus interrupt the biological pathways essential for survival of the microorganism(s).
Further growth of the on-demand chemical space size and diversity is also supported by recent development of new robust reactions for the click-like assembly of building blocks. As well as ‘classical’ azide-alkyne cycloaddition click chemistry120, recognized by the 2022 Nobel Prize in chemistry121, and optimized click-like reactions including SuFEx122, more recent developments such as Ni-electrocatalysed doubly decarboxylative cross-coupling123 show promise. Other carbon–carbon forming reactions use methyliminodiacetic acid boronates for Csp2–Csp2 couplings124, and most recently tetramethyl N-methyliminodiacetic acid boronates125 for stereospecific Csp3–C bond formation. Each of these reactions applied iteratively can generate new on-demand chemical spaces of billions of diverse compounds operating with a limited number of building blocks.
Readers are highly recommended to refer to the first edition of this chapter (4) for basic CADD concepts and classical protocols to gain a fundamental understanding about CADD methods towards antibiotics development. Pharma companies amass collections of compounds for screening in-house, whereas in-stock collections from vendors (see the figure, part a) allow fast (less than 1 week) delivery, contain unique and advanced chemical scaffolds, are easily searchable and are HTS compatible. However, the high cost of handling physical libraries, their slow linear growth, limited size and novelty constrain their applications.
At the same time, experimental testing of predictions also provides data that can feed back into improving the quality of the models by expanding their training datasets, especially for the ligand property predictions. Thus, the DL-based QSPR models will greatly benefit from further accumulating data in cell-permeability assays such as CACO-2 and MDCK, as well as new advanced technologies such as organs-on-a-chip or functional organoids to provide better estimates of ADMET and PK properties without cumbersome in vivo experiments. The ability to train ADMET and PK models with in vitro assay data representing the most relevant species for drug development (typically mouse, rat and human) would also help to address species variability as a major challenge for successful translational studies. All of this creates a virtuous cycle for improving computational models to the point at which they can drive compound selection for most DDD end points. When combined with more accurate in vitro testing, this may reduce and eventually eliminate animal test requirements (as recently indicated by FDA)145. A large number of trials are being conducted to identify binding modes of ligands and selection of the most energetically favored poses.
A binding-site identification method under the SILCS framework, named SILCS-Hotspots, was developed recently by our laboratory (115). SILCS-Hotspots is designed to identify fragment binding hotspots that are spatially distributed across the global protein structure including both surface and interior binding sites. The general protocol using SILCS-Hotspots to identify putative binding sites on a protein is described as the following. With the fast development of more powerful computing hardware, expensive algorithms such as free energy perturbation methods (23), which can only be used to finely tune the drug candidates at the lead optimization stage, become much more affordable and have been routinely used in a range of applications (24–26). Alternative CADD methods represent novel solutions that exploit the interactions between drugs and targets are also seeing wider use. Our laboratory put forward the SILCS methodology as described previously, and information from SILCS can be utilized in many different ways in various aspects of drug discovery (16–18).
The search algorithm and the scoring function are two important components for determining protein-ligand interactions [47]. The search algorithm is responsible for searching different poses and conformations of a ligand within a given target protein and the scoring function estimates the binding affinities of generated poses, ranks them, and identifies the most favorable receptor/ligand binding modes [47, 48]. An ideal search algorithm should be fast and effective, and the scoring function must be capable of determining the physicochemical properties of molecules and the thermodynamics of interactions. The existing modern drug discovery implies the knowledge of genomics, bioinformatics, computational chemistry, and combinatorial chemistry for virtual screening, HTS, Lenovo ligands. However, the concept of LBDD relies mostly on drug planning, which develops a simple drug molecule, a 3D through trial and error methods that improves the overall biological activity of the compound. Visceral Leishmaniasis (VL) is a serious public health issue, documented in more than ninety countries, where an estimated 500,000 new cases emerge each year.

Comments
Post a Comment